Abstract
Background and Aim: Azacitidine have shown clinical activity in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML), particularly at low, non-cytotoxic doses favoring hypomethylation over cytotoxicity. Cancer/testis antigens (CTAs, encoding for immunogenic proteins which are normally expressed in the testicles and trophoblastic cells of the ovary, have been shown to undergo derepression after the use of demethylating agents in cancer cell line models. We took advantage of the unique model of Aza-treated high risk MDS to in vivocharacterize candidates for immunotherapy following hypometilating therapy.
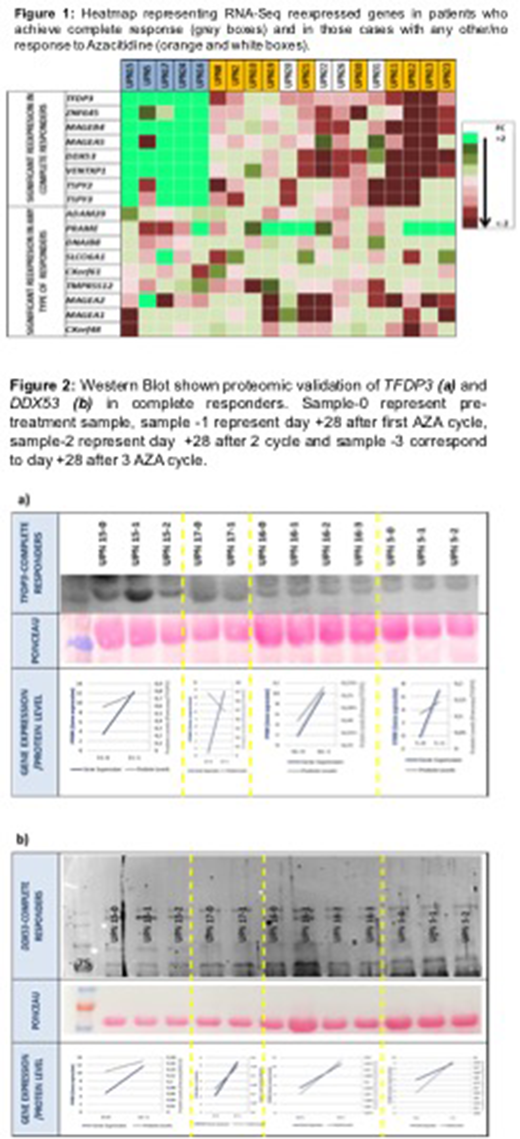
Methods:Targeted RNA-Seq (tRNA-Seq) designed to capture 214 CTA genes was performed in 19 MDS or CMML patients at day 0 and at day +28 after first AZA cycle. In 10 patients the analysis was extended to third and/or four cycles. Sequencing, coverage, mapping and alignment was performed using the Ion Torrent Browser Suite. We used LIMMA package or empirical analysis of digital gene expression data in R (edgeR) to identify differential expressed genes. A homogeneous treatment schedule with AZA (75 mg/m2/day x 7 days) was received by each patient.The response criteria were from IWG 2006, including a bone marrow and cytogenetic reevaluation after 6 cycles. The main candidates selected from de tRNA-seq experiment were characterized at protein level in cell lysate and/or plasma by Western Blot. Primary antibodies used from Thermo Fisher were: ADAM29, DDX53, PRAME, B-TUBULINA, FAM46D, B-ACT, TMPRSS12, MAGEB4, GAPDH, MAGEA1, MAGEA2, TSPY2, Cxorf48, DNAJB8, NY-ES0-1, CT83/Cxorf61 and TFDP3 from Santa Cruz Biotechnology. We used either cell lysate or plasma to study the protein levels in non-reducing SDS-PAGE at different acrylamide concentration depending on the protein size. ImageJ was used to quantify protein levels.
Results:The median age of the cohort was 69 (range 48-81 years). The cohort consisted of 16 MDS and 3 CMML patients. MDS Patients were stratified based on IPSS as low or intermediate-1 (n=14) and intermediate-2 or high (n=2) risk groups and CMML patients based on a CPSS of intermediate-2 (n=3). Five patients were classified as complete responders achieving a complete cytogenetic and/or marrow remission (CR). Regarding the tRNA-Seq, on average, we achieved 3.888.308 mapped reads per sample (p25-p75, 1.2x106 - 5.9x106) which represents a sufficient depth for digital gene expression profiling of 214 genes. CTAs re-expressed significantly after one AZA cycle in complete responders were TFDP3 (FC=6,4), ZNF645 (FC=2), MAGEB4 (FC=3,1), MAGEA5 (FC=2), DDX53 (FC=2), VENTPX1 (FC=5,5), TSPY2(FC=3,4) and TSYP3 (FC=3,5). Other potential candidates with significantly re-expression in any responders vs. non responder were:SLCO6A1, ADAM29, DDX53, PRAME, FAM46D, TMPRSS12, MAGEA1, MAGEA2, Cxorf48, DANJB8, NYESO-1 and CT83. In the western-blot experiments, protein expression, longitudinally measured in peripheral blood lysate and/or plasma showed a parallel dynamic to gene expression in the case of TFDP3and DDX53.
Conclusions:In this study, targeted RNA-seq is shown as effective and accurate tool to detect weakly expressed transcripts as CTAs. In this work, TFDP3 and DDX53, validated at proteomic level, emerge as most promising candidates for immunotherapy in combination with hypomethylating agents in MDS patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal